Table Of Content
- Scaling alt-protein production: What to know about moving from lab to commercial scale
- Graduate students
- De novo design of a hyperstable non-natural protein-ligand complex with sub-Å accuracy.
- Humans have ideas; Rosetta gives them shape
- Proteins are incredibly versatile input types, making them very attractive for deep learning training.
- How to create your protein label design
- Institute for Protein Design

Now, our group has developed a computational method called COMBS that can generate protein sequences from scratch that are able to tightly and specifically bind to challenging targets. COMBS thus enables the rapid generation of proteins that may act as sensors, therapeutics, or delivery vehicles. Aminoacyl tRNA synthetases link amino acids with their cognate tRNAs and are thus critical players in the expansion of the genetic code. However, it has proven incredibly difficult to engineer synthetases to recognize polar, non-natural amino acids that mimic common post-translational modifications (PTMs) of proteins. The ability to precisely program PTMs and monitor their effects on cell fate in disease-relevant mammalian cells has been a longstanding goal.
Scaling alt-protein production: What to know about moving from lab to commercial scale
This press release contains forward-looking statements involving risks and uncertainties, including statements regarding growth and cash flow, and PDL's actual results may differ materially from those in the forward- looking statements. As we continue to unlock the secrets of proteins and learn to harness their potential, the possibilities for protein design are virtually limitless. In this post, we will delve into the basics of protein design, explore its real-world applications, and learn about the interdisciplinary nature of this fascinating field. The synthetic cytokine neoleukin-2/15 was created in an attempt to jettison some very heavy baggage that has dogged cancer researchers for decades. Since the 1980s, the cytokine interleukin (IL)-2 has tantalized oncologists with the promise of harnessing the immune system to attack tumors. IL-2 naturally stimulates T cell proliferation, but it also exerts ferocious side effects on the body, particularly in the lungs.
Graduate students
The precise order of amino acids in a protein chain, like the letters in a word, is crucial. The unique functions of a protein stem from its three-dimensional structure, which is determined by the sequence of amino acids composing the protein. Franzi grew up in Germany and graduated from Albert Ludwigs University Freiburg with a BSc in Molecular Medicine and afterwards a MSc in Biochemistry from University of Leipzig. She pursued her PhD at the Max Planck Institute for Terrestrial Microbiology where she worked with Dr. Georg Hochberg in the area of Evolutionary Biochemistry. In her work she investigated self-assembly of proteins and the evolution of biological complexity.
De novo design of a hyperstable non-natural protein-ligand complex with sub-Å accuracy.
The world of proteins is incredibly complex, offering endless possibilities for exploration and discovery. They are found inside every living thing and act as structural components, transporters, signaling molecules, and much more. Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily. Foldit players have helped make published discoveries through their unique creativity and ingenuity. Research on it began here as an undergraduate project and would go on to form the basis of a spinout company. Algorithm DevelopmentBiomolecular modeling and computational design are at the heart of everything we do.
King created protein subunits that arrange themselves into a virus-like particles (VLPs), displaying viral antigens in regular arrays to elicit an effective immune reaction. Synthetic VLPs offer more opportunities for customization than natural ones, opening up the possibility of vaccines for intractable viruses such as RSV and HIV. King and his colleagues fused the SARS-CoV-2 spike protein to self-assembling nanoparticles and are working on creating the optimal antigen display for a coronavirus vaccine. Silva studied the 3D structure of IL-2 in its high-affinity conformation, the shape it assumes when bound to all three receptor subunits. He built a new protein that emulated that shape but was “otherwise unrelated in topology or amino acid sequence.” It also lacked any binding site for the alpha receptor subunit. By constructing a completely novel protein instead, the team could create a highly stable molecule that bound with high efficiency to the beta and gamma subunits and ignored the alpha subunit.
Proteins are incredibly versatile input types, making them very attractive for deep learning training.
Jerry is a graduate student in Harvard’s Molecules, Cells, and Organisms (MCO) program. During his gap year before starting at Harvard, he served the Taiwanese army and then worked with Dan Tawfik at the Weizmann Institute to investigate the origin of peptides and the genetic code in the origin of life. For his PhD, he hopes to elucidate the principles of biomolecular evolution using protein design, evolutionary biology, and high-throughput experimental approaches. When he is not thinking about science, he enjoys cooking, hiking, backpacking, learning languages, and exploring the evolution of human artifacts. Kevin is an undergraduate at Harvard studying computer science and molecular & cellular biology.
MICROBIAL PROTEINS AND THEIR MODIFICATIONS
Meanwhile, insulin, a hormone produced by the pancreas, helps regulate blood sugar levels by signaling cells to take in glucose from the bloodstream. Raphael's model for evaluating protein structures is posted on bioRxiv as a preprint. The protein system is called LOCKR, for “Latching, Orthogonal Cage/Key Proteins,” and it consists of a six-helix protein that folds into a stable, cage-like formation. The cage can interact with either its own ‘latch’ helix domain, which holds the cage closed, or to another protein—the ‘key’—which triggers a conformational change.

How to create your protein label design
On another level, the mission of the lab is training the next generation of scientists and leaders in the field of regenerative medicine. The lab is committed to generating the best work environment to make the best science possible, and to foster multidisciplinary research using expertise from engineering, developmental biology, medical science and computer science. Besides, they catalyze rapid chemical reactions in mild conditions such as standard pressure and non-toxic solvents. With such attractive properties, it is no wonder Biochemists and Chemists have dreamed of designing proteins with customizable properties for decades.
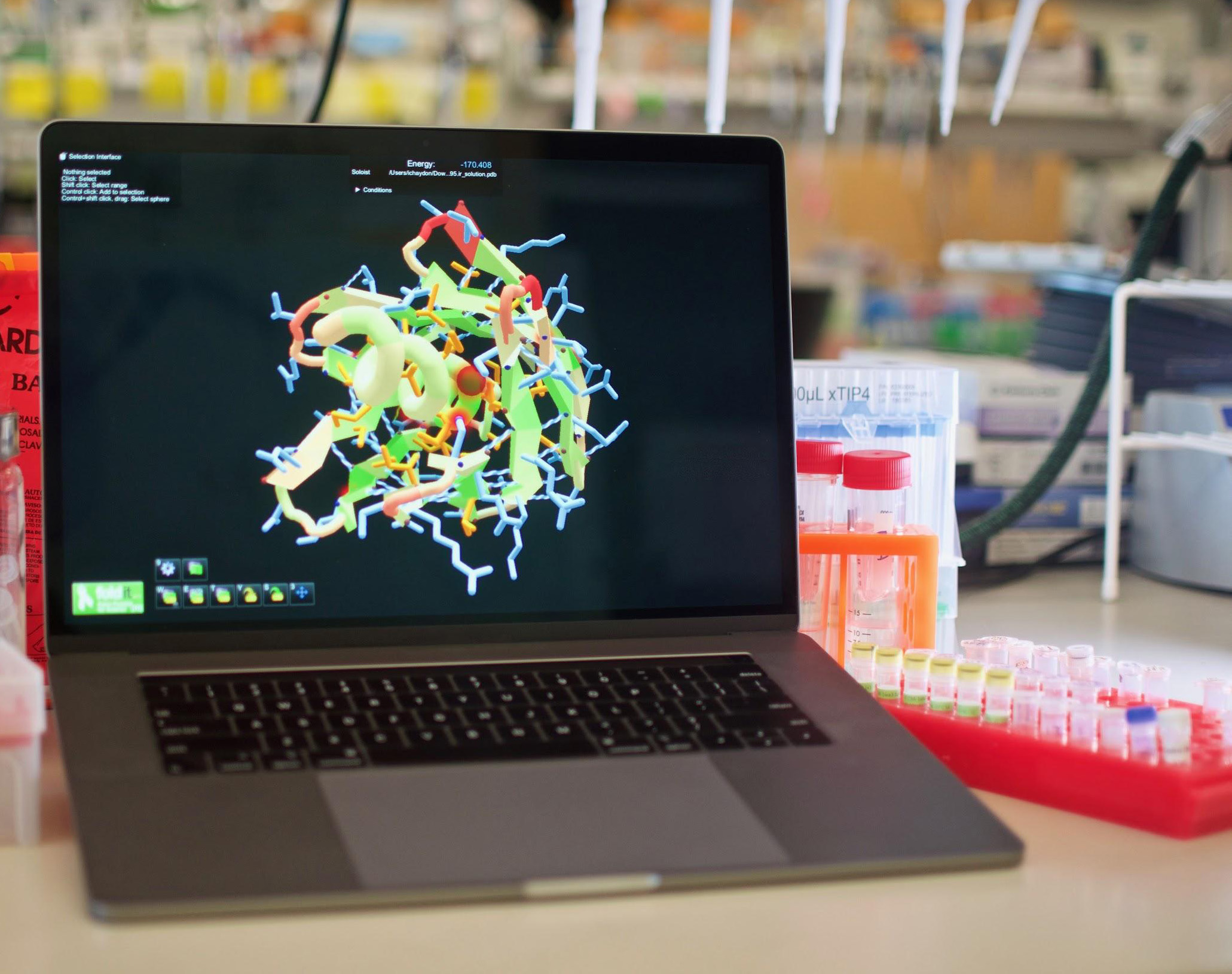
In February, Rosetta successfully predicted the 3D structure of the SARS-CoV-2 spike protein, which allows the virus to gain access to human cells. The flurry of effort that accompanied the project crashed some of the networked computers, but with the protein structure now in hand, work is underway to fight the disease. Researchers crafted mini-protein binders that perfectly complement the spike protein, preventing it from latching onto cellular receptors. To make a vaccine, IPD researchers are attaching the spike protein to a synthetic virus-like particle. The particle displays the spike protein in a repetitive array, an arrangement designed to stimulate a vigorous immune response. Researchers at the University of Washington’s Institute for Protein Design (IPD) don’t like to work alone.
The Cochran Laboratory uses interdisciplinary approaches in chemistry, engineering, and biophysics to study and manipulate complex biological systems, with a focus on developing new technologies for basic science and biomedical applications. In addition, combinatorial and rational methods are used to engineer designer protein and peptide ligands for a variety of applications including wound healing, cardiac tissue engineering, and cancer imaging and therapy. Until recently, however, the discovery of binding proteins depended on the serendipity of high-throughput experimental screening and animal inoculations.
The Rockefeller University » Jiankun Lyu - The Rockefeller University
The Rockefeller University » Jiankun Lyu.
Posted: Tue, 07 Feb 2023 17:15:21 GMT [source]
During undergrad, she worked on molecular immunology at JHU and machine learning for protein engineering at Microsoft Research. Jody is working on de novo design of ligand-binding proteins using computational and deep learning approaches. Our lab invents and uses synthetic biology tools and approaches to build tissues in the lab. Over the past 16 years, UW researchers have made significant progress in protein design and protein structure prediction, developing the world leading Rosetta software. Over this period, UW scientists have developed methods for designing proteins with a wide range of new functions, including catalysts for chemical reactions, HIV and RSV vaccine candidates, and flu virus inhibitors. The IPD integrates these strengths in protein design with Seattle-area expertise in biochemistry, engineering, computer science and medicine, and leverages the exceptional Seattle strength in the software industry.
For larger complexes involving multiple different subunits, the team was able to detect the configuration of the proteins in the complex using a mass spec technique called surface-induced dissociation. By slamming the molecules into a surface to break apart the subunits and then measuring which subunits colocalized among the resulting fragments, they could see how the subunits were arranged in the large complex. This type of structural characterization provides critical information to verify whether synthetic protein complexes are forming the way the designers expect. Today IPD lists seven ongoing projects to address the pandemic, from designing nanoparticle vaccines and anti-inflammatory proteins to screening existing drugs. Through Baker’s hypercollaborative nature and his desire to distribute the technology for broad adoption, de novo protein design may one day become a part of every protein engineer’s toolbox. Ben graduated from Johns Hopkins University with his Bachelors in Biophysics and a minor in Computer Science.
PvP was the first company launched through the University of Washington’s Translational Investigator Program. Takeda Pharmaceuticals fronted the money to conduct phase 1 clinical trials on the novel enzyme, called KumaMax, and in February 2020 exercised its option to acquire PvP. The University of Washington’s Institute for Protein Design has become a hub to galvanize de novo protein engineering, citizen science and much more since its founding in 2012. Bioactive PeptidesChemically synthesized molecules with predictable structures and functions. Advanced Drug DeliveryNanoscale protein assemblies that move therapeutics to specific cells within the body.
No comments:
Post a Comment